Projects
Define protective and pathogenic roles of LNSCs during viral infection
Thomas Morrison, PhD Publications Atlases
We hypothesize that inflammatory responses, cell trafficking, and development of protective adaptive immune responses are regulated by discrete populations of LNSCs during viral infection
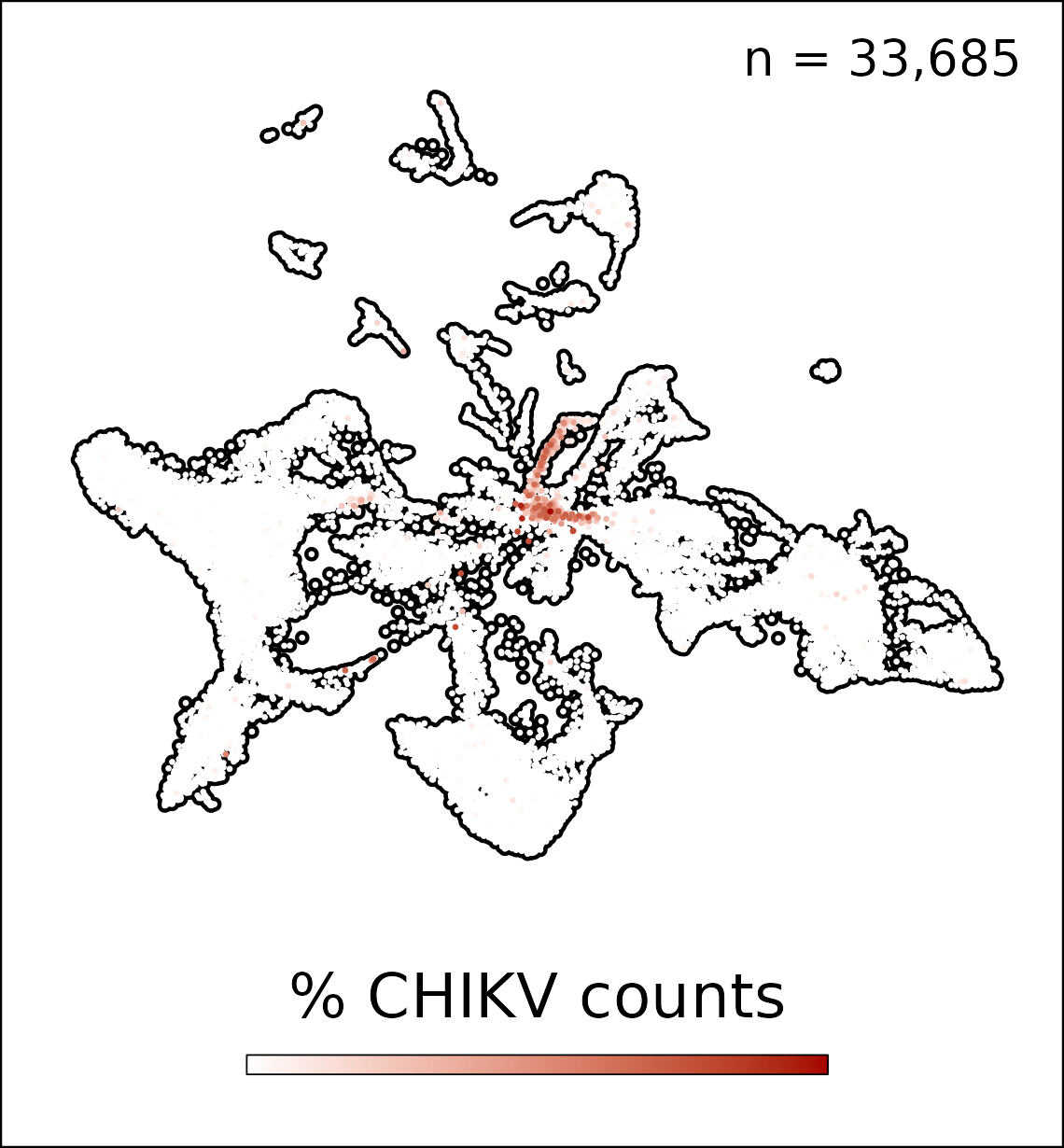 Project 1 will define how viral targeting of distinct LNSCs regulates viral dissemination, LN organization and inflammation, and development of adaptive immunity. Project 1 will establish how viral targeting of LNSCs impacts antigen archiving and immune memory to archived antigens. Finally, Project 1 will study how isotype-specific immune complexes (ICs) promote chronic viral infection of follicular dendritic cells (FDCs). This project will use novel mouse models and human tissues and advanced spatial and single-cell RNA approaches. This project will define how LNSCs shape antiviral immune responses and how viruses manipulate LNSCs to evade immunity.
Project 1 will define how viral targeting of distinct LNSCs regulates viral dissemination, LN organization and inflammation, and development of adaptive immunity. Project 1 will establish how viral targeting of LNSCs impacts antigen archiving and immune memory to archived antigens. Finally, Project 1 will study how isotype-specific immune complexes (ICs) promote chronic viral infection of follicular dendritic cells (FDCs). This project will use novel mouse models and human tissues and advanced spatial and single-cell RNA approaches. This project will define how LNSCs shape antiviral immune responses and how viruses manipulate LNSCs to evade immunity.
Identify mechanisms of vaccine antigen localization and retention by LNSCs that drive and enhance cellular and humoral immunity
Beth Tamburini, PhD Publications Atlases
We hypothesize that LNSC targeting improves and extends the cellular and humoral responses required for vaccine mediated immunity
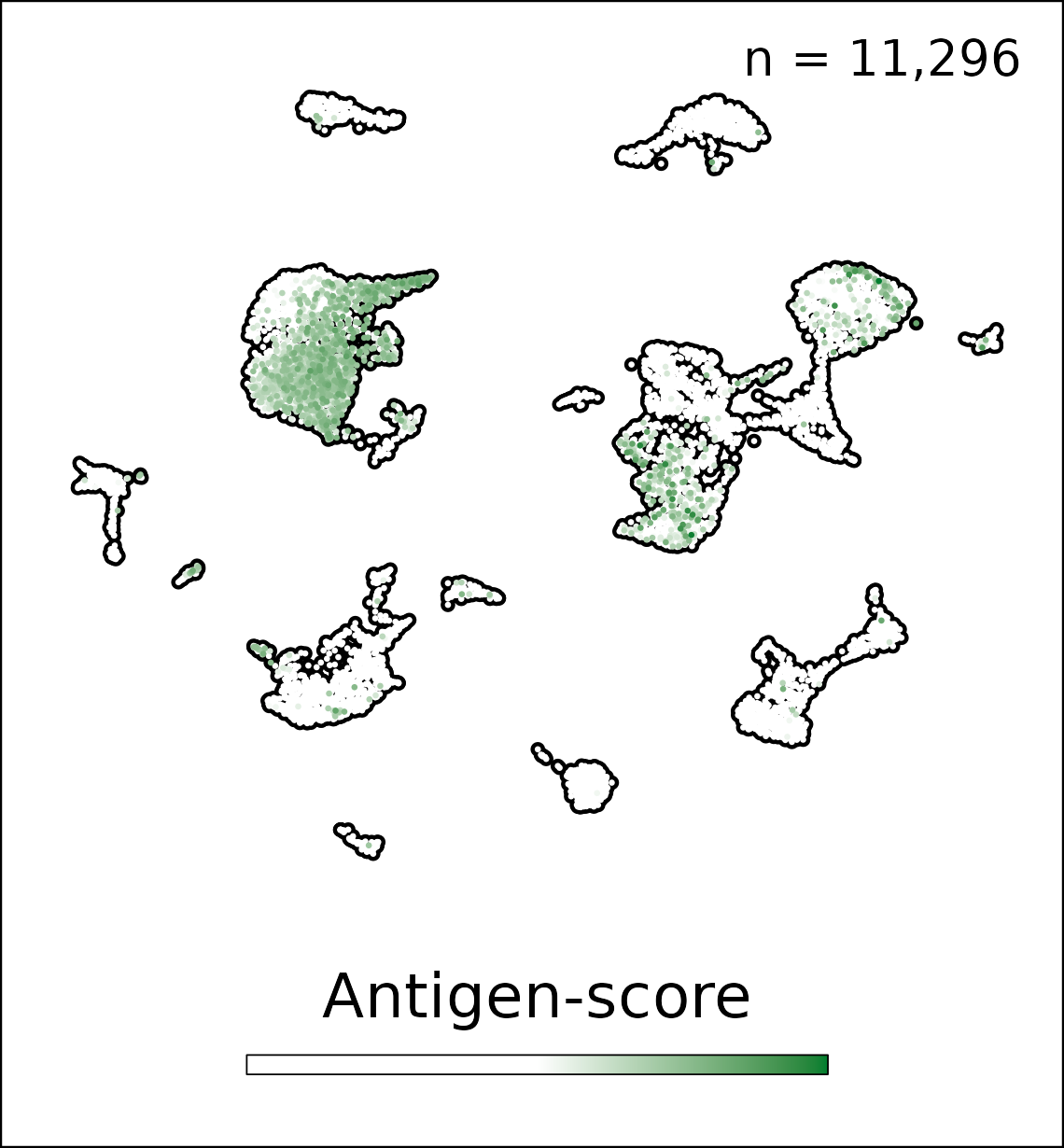 Project 2 will define mechanisms of vaccine antigen acquisition and retention by different LNSCs. Project 2 will determine if manipulation of antigen duration in LNSCs improves vaccine protection against arboviruses and influenza virus. Finally, Project 2 will define the consequences of virus induced cell death on LEC ‘training’ and the impact on vaccine immunity. This project will distinguish functions of specific LNSC populations and create novel vaccine strategies (e.g., mRNA, virus-like particles, and lipid nanoparticles) and apply sophisticated spatial and single-cell transcriptomic approaches to evaluate antigen levels within infrequent and transcriptionally defined LNSC subsets. This project will determine how specific LNSC subsets shape vaccine immunity and the consequences of viral infection of LNSCs to vaccine immunity.
Project 2 will define mechanisms of vaccine antigen acquisition and retention by different LNSCs. Project 2 will determine if manipulation of antigen duration in LNSCs improves vaccine protection against arboviruses and influenza virus. Finally, Project 2 will define the consequences of virus induced cell death on LEC ‘training’ and the impact on vaccine immunity. This project will distinguish functions of specific LNSC populations and create novel vaccine strategies (e.g., mRNA, virus-like particles, and lipid nanoparticles) and apply sophisticated spatial and single-cell transcriptomic approaches to evaluate antigen levels within infrequent and transcriptionally defined LNSC subsets. This project will determine how specific LNSC subsets shape vaccine immunity and the consequences of viral infection of LNSCs to vaccine immunity.
Cell and molecular mechanisms of antigen archiving by lymphatic endothelial cells
Jay Hesselberth, PhD Publications Atlases
We hypothesize that antigen archiving is achieved by stable compartmentalization of acquired material in endosomes.
 Project 3 will define cellular and molecular mechanisms promoting stable retention of protein and RNA in LECs. Project 3 will determine how LECs stably retain ntigens and how this process is regulated by compartmentalization in endosomes. This project will develop and use innovative probes of cellular localization in vivo, to evaluate antigen levels within LECs and immune cells.
Project 3 will define cellular and molecular mechanisms promoting stable retention of protein and RNA in LECs. Project 3 will determine how LECs stably retain ntigens and how this process is regulated by compartmentalization in endosomes. This project will develop and use innovative probes of cellular localization in vivo, to evaluate antigen levels within LECs and immune cells.
Cores
Mouse transgenics and tissue characterization
Jennifer Matsuda, PhD Publications
Core B will generate novel genetic mouse models and develop a tissue and cell repository, this includes the following tasks.
Task 1. Generation of novel mouse lines
Task 2. Provide a mouse colony for LNSC biology
Task 3. Provide a resource for maintenance and distribution of human and mouse LNSCs and lymphoid tissuesMolecular technologies and informatics
Jay Hesselberth, PhD Ryan Sheridan, PhD Publications Atlases
Core C will apply new molecular tools and bioinformatic approaches to understand the contributions of lymph node stromal cells to immune responses, this includes the following tasks.
Task 1. Generate and supply validated reagents for single-cell and spatial transcriptomic experiments to evaluate LNSC contributions to immunity and infection
Task 2. Provide a bioinformatics resource for rigorous and reproducible analysis of sequencing experiments
Task 3. Develop pipelines for large scale meta-analysis of single-cell and spatial transcriptomic data